SANTÉ

G4_ G quadruplexes


G4_ G quadruplexes
Structure des Acides Nucléiques, Télomères et Évolution
Les télomères sont des complexes nucléoprotéiques qui protègent les extrémités des chromosomes linéaires des eucaryotes. Les recherches de l’équipe SANTE portent sur la régulation des télomères, tant dans des contextes normaux que pathologiques, ainsi que sur l’évolution des télomères.
Au-delà des télomères, l’arrivée en 2022 d’un chimiste au sein de l’équipe a ouvert de nouvelles perspectives de recherche dans le domaine de la chimie organique.
1 - Structures non-canoniques des acides nucléiques et interactions avec des protéines de liaison à l’ADN simple-brin
2 - Évolution des télomères
3 - Dysfonctions télomériques
4 - Amélioration des systèmes rapporteurs bioluminescents
1 - Interaction des protéines de liaison à l’ADN simple-brin impliquées dans le maintien des télomères avec les structures secondaires de l’ADN télomérique
2 - Nouvelles approches pour interférer avec la régulation des télomères : épigénétique, petites molécules de synthèse et AsiDNA
3 - Recherche de protéines interagissant avec les structures G4

Equipe Santé 2025
• Carole Saintomé, PR Sorbonne Université J’ai obtenu mon doctorat en chimie et biochimie à l’Université Pierre-Marie Curie (aujourd’hui Sorbonne Université) à Paris et j’ai rejoint le laboratoire en 2011. Je m’intéresse à la compréhension du mécanisme moléculaire qui conduit au maintien des télomères, dans des contextes normaux et pathologiques, ainsi que durant l’évolution. Mon axe de recherche porte sur l’investigation des interactions entre les protéines liant l’ADN simple brin et les acides nucléiques à travers une approche multidisciplinaire à l’intersection de la chimie et de la biochimie.
• Patrizia Alberti, MC Muséum J’ai obtenu mon diplôme en physique à l’Université de Milan et mon doctorat en biophysique au Muséum National d’Histoire Naturelle à Paris. Après un post-doctorat dans une entreprise de biotechnologie, j’ai rejoint ce laboratoire en 2005. Mon principal axe de recherche est la caractérisation des structures non canonique des acides nucléiques et la manière dont les protéines interagissent avec elles. Je m’intéresse particulièrement aux séquences répétées et à l’évolution des télomères, qui sont une illustration fascinante de l’unité et de la diversité de la vie.
• Jian-Sheng Sun, PR Muséum J’ai obtenu un doctorat en biophysique à l’Université Pierre-Marie Curie (aujourd’hui Sorbonne Université) à Paris. J’ai mené une double carrière : dans le secteur académique (CNRS/MNHN, 25 ans) en étudiant les structures et fonctions des acides nucléiques, les dommages et réparations de l’ADN, et dans la biotechnologie (co-fondateur, PDG et président de DNA Therapeutics, 10 ans) pour développer un médicament anticancéreux de première classe constitué d’ADN double brin, en intervenant sur les mécanismes de signalisation des dommages à l’ADN. Actuellement au MNHN, je dirige le département des « Adaptations des organismes vivants ». Je suis également le directeur scientifique du « Bioinspire-Muséum », le co-pilote de l’ »Approche globale de la santé » des programmes SOUND de l’Alliance des Universités de la Sorbonne et expert scientifique impliqué dans les négociations sur le CBD-DSI durant la présidence française de l’Union européenne et les réunions subséquentes du CBD-COP15
• Anne-Laure Guieysse-Peugeot, PR Muséum Pr. Anne-Laure Guieysse-Peugeot a obtenu son doctorat en biologie moléculaire au Muséum National d’Histoire Naturelle à Paris. Tout au long de sa carrière au Muséum, Anne-Laure Guieysse-Peugeot a contribué à la recherche sur les mécanismes de régulation et l’expression génique. Ce travail a commencé par l’étude de la structure en triple hélice de l’ADN et de son implication dans la régulation de l’expression génique (1991-2000). La découverte des mécanismes épigénétiques et leur rôle dans le contrôle de l’expression génique l’ont amenée à explorer le rôle de la méthylation de l’ADN dans la régulation transcriptionnelle (2004-2014), suivie par l’étude des rôles des mécanismes épigénétiques dans la régulation des télomères durant le processus de vieillissement (2014-présent)
• Laureline Roger, MC Muséum J’ai obtenu mon doctorat en biologie moléculaire et cellulaire à l’Université de Montpellier (France) et j’ai ensuite effectué mon postdoctorat à la Cardiff School of Medicine (Pays de Galles, Royaume-Uni) dans le laboratoire de Duncan Baird, où j’ai étudié le rôle de la dysfonction des télomères dans la carcinogenèse. Pendant cette période, j’ai également collaboré avec Kristin Ladell, où j’ai utilisé l’analyse de la longueur des télomères uniques (STELA) pour étudier l’historique réplicatif et la différenciation des cellules T mémoire. J’ai rejoint le Muséum National d’Histoire Naturelle en tant que chargé de cours en 2014. Mes recherches actuelles se concentrent sur l’identification des déclencheurs et des mécanismes des fusions de télomères et sur la façon dont la dysfonction des télomères peut contribuer à l’initiation du cancer.
• Yves Janin, DR CNRS Le Dr Yves L. Janin a obtenu un doctorat en chimie organique en 1993 sous la direction du Dr Émile Bisagni à l’Institut Curie. Après deux ans de postdoctorat à l’ICSN de Gif/Yvette et une année à la Danish School of Pharmacy à Copenhague, il a ensuite travaillé six ans à l’Institut Curie. Après un congé sabbatique dans les laboratoires Aventis à Vitry-sur-Seine, il a rejoint l’Institut Pasteur jusqu’en 2022. Il travaille maintenant au Muséum National d’Histoire Naturelle à Paris. Ses intérêts portent sur la chimie médicinale, les nouvelles entités chimiques, ainsi que sur la conception de systèmes de reporting originaux basés sur la bioluminescence.
• Gildas Mouta Cardoso, IE Muséum J’ai rejoint le MNHN en 2003 pour travailler sur une plateforme de protéomique. Après avoir obtenu un Master en chimie en 2011, j’ai intégré l’équipe Santé en tant que chimiste organique pour synthétiser des ligands de G-quadruplexes. Mon principal axe de recherche est la synthèse de petites molécules pour les projets développés par l’équipe. Je participe également à des projets avec d’autres équipes du laboratoire
• Drayton- Libotte Bernadette, IE INSERM
• Virginie Hossard, AI Inserm J’ai obtenu un BTS en biochimie en 2005 et un poste d’assistante ingénieure en expérimentation animale en 2007 au sein d’une équipe travaillant sur les rythmes biologiques et les cancers à l’Hôpital Paul Brousse. J’ai rejoint le laboratoire en 2015 pour travailler sur plusieurs sujets en biologie cellulaire et moléculaire (ligands G4, ADN mitochondrial, ARN TERRA, C-Circles) et plus récemment en biochimie sur la caractérisation des protéines POT1 et RPA impliquées dans la stabilité télomérique
• Marianne Bechara, Doctorante MNHN Après avoir obtenu un diplôme de pharmacie à l’Université Saint-Joseph au Liban, j’ai poursuivi un master en génétique et épigénétique à l’Université de la Sorbonne. J’ai rejoint le laboratoire en janvier 2023 en tant que stagiaire, et je suis actuellement en première année de doctorat
• Inès Salhi, Doctorante MNHN
• Pavlina Thermidi, Doctorante CNRS
• Nina Amrane, L3 apprentissage
• Patrick Mailliet, Chercheur Bénévole Muséum J’ai passé environ 40 ans dans l’industrie pharmaceutique en tant que chef d’équipe en chimie médicinale et chef de projet en oncologie, avec une expertise spécifique dans les composés interagissant avec l’ADN et les kinases. Depuis 2010, je travaille au MNHN sur des ligands de structures d’acides nucléiques (principalement des G-quadruplexes). J’apporte également des conseils à l’équipe sur les avancées en chimie médicinale et j’enseigne dans différents masters. En plus de ces activités, je suis consultant pour deux petites entreprises pharmaceutiques, une en France et une en Italie
Anciens membres 2014 - 2025
Congcong Li, Doctorante Muséum (2022-2025) Jean-François Riou, PR Muséum (2007-2023)
Anthony Bugaut, CR CNRS (2013-2020)
François Peurois, ATER (2018-2019)
Coralie Modeste, IE (2018-2019)
Florian Gourmelon, IE (2022-2024)
Jean Chatain, PhD student (2017-2020)
Gabriel Le Berre, PhD student (2016-2019)
Pauline Lejault, PhD student (2014-2017)
Frédéric Thiebaut, PhD student (2014-2017)
Illustrations des différents thèmes de l’équipe

Droplets formed by human RPA under fluorescent microscopy

G4 at telomeres

Walking along telomeres: G4, hairpins and SSB proteins

Interfering with telomeres: epigenetic and ligands
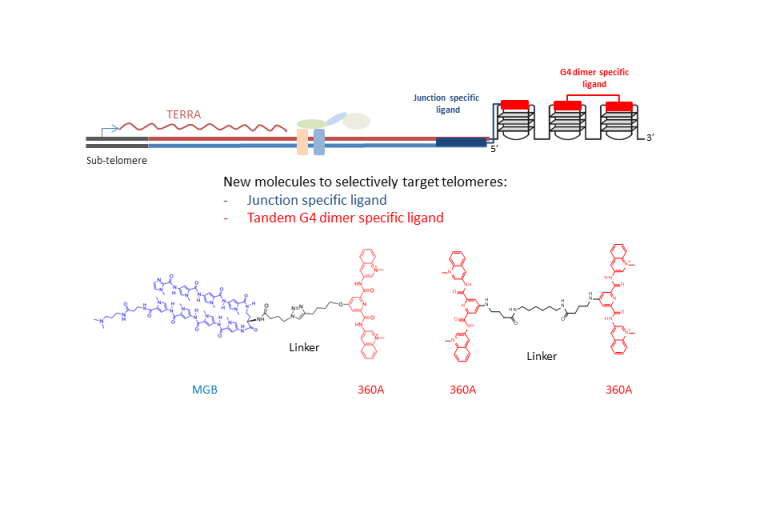
New molecules specifically targeting telomeres

Interfering with telomeres: AsiDNA

RPA binding with multimeric telomeric G-quadruplexes studied by EMSA

Single TElomere Length Analysis (STELA)

Telomere crisis
Nous possédons une expertise dans la caractérisation des structures non-canoniques de l’ADN et de leur interaction avec des protéines de liaison à l’ADN simple-brin. Nous nous intéressons particulièrement aux séquences d’ADN composées de répétitions en tandem de 2 à 6 nucléotides, également connues sous le nom de STR (pour « Short Tandem Repeats »).
Notre objectif est de comprendre la plasticité et l’évolution des eucaryotes au niveau des structures moléculaires qui assurent la protection des extrémités de leurs chromosomes.
Chez les eucaryotes où les télomères sont allongés par la télomérase, le brin d’ADN télomérique orienté vers l’extrémité 3› est généralement composé de répétitions d’un court motif (dit « motif télomérique ») contenant des guanines consécutives. L’ADN télomérique s’associe (directement ou indirectement) à un ensemble de protéines spécifiques des télomères, qui sont essentielles à la protection et régulation des télomères.
Notre objectif est de comprendre comment certaines protéines (forme sauvage, variante ou isoforme) participent à la stabilité ou à l’instabilité des télomères.
Ces dernières années, nous avons étudié comment hRPA (Replication Protein A humaine) et hPOT1-TPP1 (deux complexes de liaison à l’ADN simple-brin impliqués dans le maintien des télomères) interagissent avec les structures secondaires (G-quadruplexes) formées par l’ADN télomérique. Dans le cadre d’un projet de recherche collaboratif financé par l’ANR (TeloRPA), nous avons lancé un nouvel axe de recherche visant à caractériser les variantes de hRPA et hPOT1 identifiées chez des patients atteints de troubles liés à la biologie des télomères (TBD pour « telomeres biology disorders »).
L’un de nos projets de recherche porte sur l’étude des systèmes rapporteurs basés sur la bioluminescence, qui sont composés d’une luciférine et d’une luciférase. Comme illustré ci-dessous, la coelentérazine est le substrat d’une large gamme de luciférases marines. Sa décarboxylation oxydative conduit à la production de coelentéramide, accompagnée de l’émission d’un photon bleu, la couleur la plus visible sous la mer. Nos travaux antérieurs ont conduit à un processus de synthèse pour préparer des luciférines O-acétylées, telles que les hikarazines-103 et 108 (doi: 10.1039/c9ob00459a; doi: 10.1002/chem.201904844).



Chez les eucaryotes où les télomères sont allongés par la télomérase, le brin d’ADN télomérique orienté vers l’extrémité 3› (« brin G ») est généralement composé de répétitions d’un court motif de 5 à 8 nucléotides (« motif télomérique ») contenant 2, 3 ou 4 guanines consécutives. Chez de nombreux eucaryotes, ce brin se termine par une extrémité 3› simple-brin. Le motif télomérique humain est l’hexamère GGGTTA. Ce motif a d’abord été identifié dans les télomères humains ; il est conservé chez les vertébrés et se retrouve chez de nombreux autres eucaryotes. La présence de guanines consécutives rend le brin G télomérique susceptible de se replier en quadruplexes de guanines (G-quadruplexes ou G4) (doi: 10.1093/nar/gkq1292).
Interférer avec la régulation des télomères présente un double intérêt : d’une part, cela permet de mieux comprendre la régulation des télomères, et d’autre part, cela constitue une approche potentielle contre la prolifération des cellules cancéreuses. La figure ci-dessous illustre les résultats majeurs de nos études.
Dans le cadre d’un projet collaboratif financé par l’ANR (G4-TopIPro), nous avons utilisé des structures G4 chimiquement contraintes pour stabiliser une topologie particulière d’ADN ou d’ARN en G4, afin de les utiliser comme appâts pour identifier des protéines interagissant avec les G4. Cela nous a permis d’identifier de nouvelles protéines interagissant avec les G4, comme le complexe NELF, impliqué dans le mécanisme de pause de l’ARN Pol II. Le complexe NELF et la pause de l’ARN Pol II favorisent l’induction de cassures double brin de l’ADN après stabilisation des G4 par la Pyridostatine (doi: 10.1038/s41598-021-92806-8).